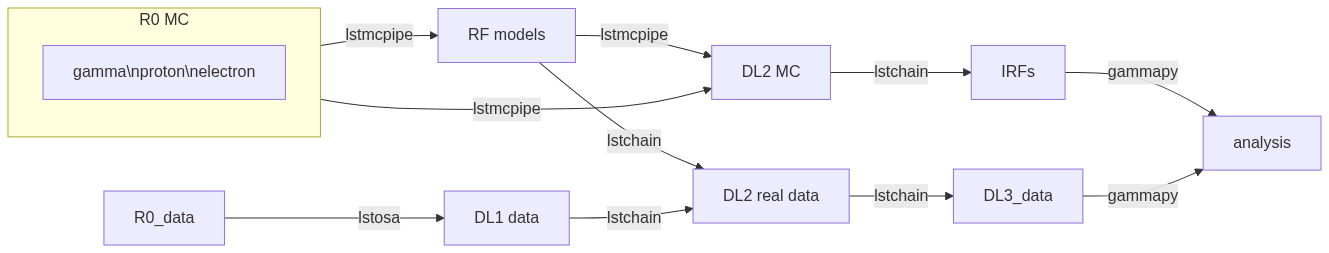
Analyzers documentation#
Context: LST data analysis#
An example of workflow:
Each step of the analysis is implemented in lstchain as a script.
For example, if you want to train a model, you may run lstchain_mc_train_pipe.
See https://cta-observatory.github.io/cta-lstchain/introduction.html#analysis-steps
Problems#
lstchain provides individual steps for each stage of the analysis but no complete pipeline
theses steps need to be orchestrated (dependencies, inputs/outputs)
data specific analysis
MC are tuned from DL1 level to match real data to be analyzed
therefore, the analysis of MC data models productions of models/IRFs are in the hands of analyzers
job handling can be harder than it seems (logic between them, directory organization, jobs requirements for different configs…)
everybody makes mistakes, except Abelardo
How do we allow custom MC productions for specific analysis in an easy-to-use and centralized manner?#
a common implementation of the pipeline
a centralized production library
a way to request for specific productions
=> lstmcpipe
General Idea#
common implementation of a pipeline such as
customizable through config file
allowing complete reproducibility of the steps
Advantages for analyzers#
easy to use (no or minimal code to write)
centralized production library
reproducibility
easy to share with other analyzers and LST Collaboration
less time, errors and frustration
Advantages for the LST Collaboration#
centralized production library
common implementation
increased trust
results are easier to compare / reproduce
less computing resources
Implementation#
lstmcpipe config file example#
workflow_kind: lstchain # should not be modified for LST analyses
prod_id: 20230517_v0.9.13_large_offset # you define this !
source_environment:
source_file: /fefs/aswg/software/conda/etc/profile.d/conda.sh # do not modify unless you have your own conda installation (and have a good reason for it)
conda_env: lstchain-v0.9.13 # name of the conda environment to use for the analysis (must exist on the cluster)
slurm_config:
user_account: dpps # slurm user account to use. Keep dpps if the production is run through the PR scheme
stages_to_run: # list of stages to run
- r0_to_dl1
- merge_dl1
- train_pipe
# all stages have a list of input/output (mandatory), lstchain options and slurm options
stages:
r0_to_dl1:
- input:
/fefs/aswg/data/mc/DL0/LSTProd2/TrainingDataset/GammaDiffuse/dec_931/sim_telarray/node_corsika_theta_69.809_az_90.303_/output_v1.4/
output:
/fefs/aswg/data/mc/DL1/AllSky/20240131_allsky_v0.10.5_all_dec_base/TrainingDataset/dec_931/GammaDiffuse/node_corsika_theta_69.809_az_90.303_
- input:
/fefs/aswg/data/mc/DL0/LSTProd2/TrainingDataset/GammaDiffuse/dec_931/sim_telarray/node_corsika_theta_69.809_az_269.697_/output_v1.4/
output:
/fefs/aswg/data/mc/DL1/AllSky/20240131_allsky_v0.10.5_all_dec_base/TrainingDataset/dec_931/GammaDiffuse/node_corsika_theta_69.809_az_269.697_
merge_dl1:
- input:
/fefs/aswg/data/mc/DL1/AllSky/20230927_v0.10.4_crab_tuned/TrainingDataset/dec_2276/GammaDiffuse
output:
/fefs/aswg/data/mc/DL1/AllSky/20230927_v0.10.4_crab_tuned/TrainingDataset/dec_2276/GammaDiffuse/dl1_20230927_v0.10.4_crab_tuned_dec_2276_GammaDiffuse_merged.h5
options: --pattern */*.h5 --no-image
extra_slurm_options:
partition: long
train_pipe:
- input:
gamma:
/fefs/aswg/data/mc/DL1/AllSky/20240131_allsky_v0.10.5_all_dec_base/TrainingDataset/dec_931/GammaDiffuse/dl1_20240131_allsky_v0.10.5_all_dec_base_dec_931_GammaDiffuse_merged.h5
proton:
/fefs/aswg/data/mc/DL1/AllSky/20240131_allsky_v0.10.5_all_dec_base/TrainingDataset/dec_931/Protons/dl1_20240131_allsky_v0.10.5_all_dec_base_dec_931_Protons_merged.h5
output: /fefs/aswg/data/models/AllSky/20240131_allsky_v0.10.5_all_dec_base/dec_931
extra_slurm_options:
partition: xxl
mem: 100G
cpus-per-task: 16
# the following stages, even if defined, will not be run as they are not in stages_to_run
dl1_to_dl2:
...
dl2_to_irfs:
...
How it works#
Each stage is a step of the pipeline and runs a lstchain script
e.g.
r0_to_dl1runslstchain_mc_r0_to_dl1
Each stage takes a list of input/output (mandatory)
Other options can be passed to the lstchain script through the config file
Slurm job options can be passed to each stage using
extra_slurm_optionslstmcpipe implements the logic between the stages and the corresponding slurm rules
e.g. waiting for all jobs from
r0_to_dl1to be over before running stagemerge_dl1
Generating a config file#
The config file can be created or modified manually. So you can define your own pipeline quite easily, use your own conda environment and target your own directories…
But can be more convienently generated from the command line lstmcpipe-generate-config that must be run on the cluster
Pipelines and paths handling#
When generating a config, lstmcpipe builds the directory structure for you.
For example, it knows that
R0data for the allsky prod are stored in/fefs/aswg/data/mc/DL0/LSTProd2/TrainingDataset/Protons/dec_*and will create the subsequent directory structure, producing DL1 data in/fefs/aswg/data/mc/DL1/AllSky/$PROD_ID/TrainingDataset/dec_*
That knowledge is implemented in the
lstmcpipe.config.paths_config.PathConfigchild classes (one child class per pipeline).The main pipelines are implemented.
You can implement your own if you have specific use cases.
The class name is passed to the
lstmcpipe-generate-configcommand line tool, along with options specific to that class.e.g.
lstmcpipe_generate_config PathConfigAllSkyFull --prod_id whatagreatprod --dec_list dec_2276you may find the supported pipelines and their generation command-line in the lstmcpipe documentation
lstchain config#
The command line tool lstmcpipe-generate-config also generates a lstchain config file for you.
It actually uses lstchain_dump_config --mc to dump the lstchain config from the version installed in your current environment. The config file is:
- complete
tailored to MC analysis
You should modify it to your needs, e.g. adding the parameters provided by lstchain_tune_nsb.
Even though lstchain does not strictly require an exhaustive config, please provide one. It will help others and provide a more explicit provenance information.
📊 Requesting a MC analysis#
As a LST member, you may require a MC analysis with a specific configuration, for example to later analyse a specific source with tuned MC parameters.
Production list#
You may find the list of existing productions in the lstmcpipe documentation. Please check in this list that a request similar to the one you are about to make does not exist already!
Determine your needs#
Depending on the real data you want to analyse.
=> determine the corresponding MC data (e.g. which training declination line ).
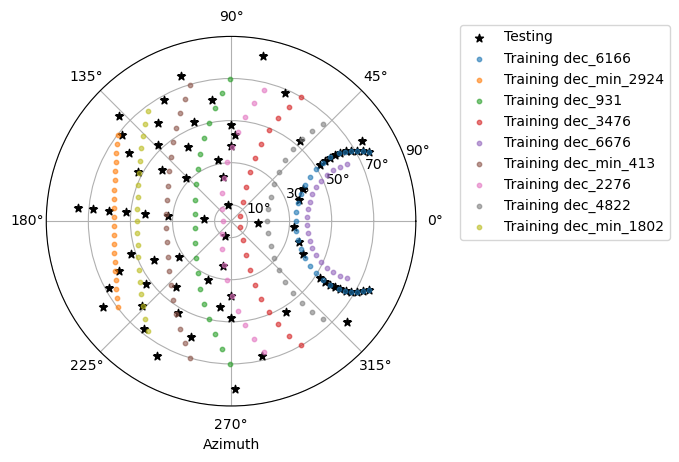
You may also check existing ones in /fefs/aswg/data/mc/DL0/LSTProd2/TrainingDataset/
=> determine if you need a tuned MC production.
Generate your config#
For most analyzers, the easiest way is to use an official conda environment on the cluster and run the lstmcpipe_generate_config command line tool.
For the allsky prod, you may use the following command line:
lstmcpipe_generate_config PathConfigAllSkyFull --prod_id whatagreatprod --dec_list dec_2276
your prod_id should be unique and explicit. It will be used to name the directories and files of your production. Add the date and the lstchain version to it, e.g.
20240101_v0.10.4_dec_123_crab_tunedthen edit the lstmcpipe config file, especially for the conda environment that you want to use for the analysis.
check that the rest of the config is ok for you (stages, directories…)
edit the lstchain config file, especially to add any NSB tuning parameters. Please provide an exhaustive config that will help others and provide a more explicit provenance information.
see
lstchain_tune_nsbfor more information
Prepare your pull-request#
If you are not familiar with git, I recommend you follow the git course: https://escape2020.github.io/school2021/posts/clase04/
make sure you are in the GitHub cta-observatory organization - if not ask Karl Kosack or Max Linhoff to add you.
make sure you are in the GitHub lst-dev group - if not ask Thomas Vuillaume, Ruben Lopez-Coto, Abelardo Moralejo or Max Linhoff to add you.
clone the lstmcpipe repository
git clone git@github.com:cta-observatory/lstmcpipe.git
(
git clone https://github.com/cta-observatory/lstmcpipe.gitif you did not setup ssh key in your github account)
create a directory named after your
prod_idinlstmcpipe/production_configs/add your lstmcpipe config, lstchain config and a readme.md file in this directory
commit and push your changes in a new branch
git switch -c my_new_branch
git add production_configs/my_prod_id
git commit -m "my new production"
git push origin my_new_branch
go to the lstmcpipe repository and create a pull-request from your branch
wait for the CI to run and check that everything is ok
your pull-request will be reviewed and merged if everything is ok
you will get notified in the pull-request when the production is ready
And then?#
We will run the production on the cluster using the lstanalyzer account with:
lstmcpipe -c lstmcpipe_config.yml -conf_lst lstchain_config.json
And will notify you in the github pull-request when it is done.
NB: standard users do not have the writing rights on /fefs/aswg/data/ so you will not be able to run the production yourself.
🚀 TL;DR - Summary for analyzers in a hurry#
Search the production library for an existing one that suits your needs
If you find one, you can use it directly (models and DL2 paths are in the config file)
If not, you can generate a config file for your own analysis. See lstmcpipe documentation for the list of supported pipelines. Example:
ssh cp02
source /fefs/aswg/software/conda/etc/profile.d/conda.sh
conda activate lstchain-v0.10.5
cd lstmcpipe/production_configs
mkdir 20240101_my_prod_id; cd 20240101_my_prod_id;
lstmcpipe_generate_config PathConfigAllSkyFull --prod_id 20240101_my_prod_id --dec_list dec_2276
Check the generated lstmcpipe config. In particular for the conda environment that you want to use for the analysis.
Edit the generated lstchain config. In particular to add any NSB tuning parameters. Please provide an exhaustive config that will help others and provide a more explicit provenance information.
Create and edit a
readme.mdfile in the same directory to describe your production. Don’t add sensitive or private information in this file, refer to the LST wiki if needed.Submit your configs+readme through a pull-request (see dedicated section) in the lstmcpipe repository.
Enjoy! 🎉 😎
Need help?#
Find this documentation again at https://cta-observatory.github.io/lstmcpipe/
Join the CTA North slack and ask for help in the lstmcpipe_prods channel
Cite lstmcpipe#
If you publish results / analysis, please consider citing lstmcpipe:
https://cta-observatory.github.io/lstmcpipe/index.html#cite-us
@misc{garcia2022lstmcpipe,
title={The lstMCpipe library},
author={Enrique Garcia and Thomas Vuillaume and Lukas Nickel},
year={2022},
eprint={2212.00120},
archivePrefix={arXiv},
primaryClass={astro-ph.IM}
}
in addition to the exact lstmcpipe version used from
You may also want to include the config file with your published code for reproducibility 🔄
Appendix#
Steps explanation 🔍#
The directory structure and the stages to run are determined by the config stages.
After that, the job dependency between stages is done automatically.
- If the full workflow is launched, directories will not be verified as containing data. Overwriting will only happen when a MC prods sharing the same prod_id and analysed the same day is run
- If each step is launched independently (advanced users), no overwriting directory will take place prior confirmation from the user
Example of default directory structure for a prod5 MC prod:#
/fefs/aswg/data/
├── mc/
| ├── DL0/20200629_prod5_trans_80/{particle}/zenith_20deg/south_pointing/
| | └── simtel files
| |
| ├── running_analysis/20200629_prod5_trans_80/{particle}/zenith_20deg/south_pointing/
| | └── YYYYMMDD_v{lstchain}_{prod_id}/
| | └── temporary dir for r0_to_dl1 + merging stages
| |
| ├── analysis_logs/20200629_prod5_trans_80/{particle}/zenith_20deg/south_pointing/
| | └── YYYYMMDD_v{lstchain}_{prod_id}/
| | ├── file_lists_training/
| | ├── file_lists_testing/
| | └── job_logs/
| |
| ├── DL1/20200629_prod5_trans_80/{particle}/zenith_20deg/south_pointing/
| | └── YYYYMMDD_v{lstchain}_{prod_id}/
| | ├── dl1 files
| | ├── training/
| | └── testing/
| |
| ├── DL2/20200629_prod5_trans_80/{particle}/zenith_20deg/south_pointing/
| | └── YYYYMMDD_v{lstchain}_{prod_id}/
| | └── dl2 files
| |
| └── IRF/20200629_prod5_trans_80/zenith_20deg/south_pointing/
| └── YYYYMMDD_v{lstchain}_{prod_id}/
| ├── off0.0deg/
| ├── off0.4deg/
| └── diffuse/
|
└── models/
└── 20200629_prod5_trans_80/zenith_20deg/south_pointing/
└── YYYYMMDD_v{lstchain}_{prod_id}/
├── reg_energy.sav
├── reg_disp_vector.sav
└── cls_gh.sav
DL0 directory structure for allsky prod#
/fefs/aswg/data/mc/DL0/LSTProd2/TrainingDataset/
├── GammaDiffuse
│ ├── dec_2276
│ ├── dec_3476
│ ├── dec_4822
│ ├── dec_5573
│ ├── dec_6166
│ ├── dec_6166_high_density
│ ├── dec_6676
│ ├── dec_931
│ ├── dec_min_1802
│ ├── dec_min_2924
│ └── dec_min_413
└── Protons
├── dec_2276
├── dec_3476
├── dec_4822
├── dec_6166
├── dec_6166_high_density
├── dec_6676
├── dec_931
├── dec_min_1802
├── dec_min_2924
└── dec_min_413
Example DL1 directory structure#
/fefs/aswg/data/mc/DL1/AllSky/20240131_allsky_v0.10.5_all_dec_base/
├── TestingDataset
│ ├── dl1_20240131_allsky_v0.10.5_all_dec_base_node_theta_10.0_az_102.199__merged.h5
│ ├── dl1_20240131_allsky_v0.10.5_all_dec_base_node_theta_10.0_az_248.117__merged.h5
│ ...
│ ├── dl1_20240131_allsky_v0.10.5_all_dec_base_node_theta_82.155_az_271.199__merged.h5
│ ├── dl1_20240131_allsky_v0.10.5_all_dec_base_node_theta_82.155_az_79.122__merged.h5
│ ├── node_theta_10.0_az_102.199_
│ ├── node_theta_10.0_az_248.117_
│ ├── node_theta_14.984_az_175.158_
│ ├── node_theta_14.984_az_355.158_
│ ...
│ └── node_theta_82.155_az_79.122_
└── TrainingDataset
├── dec_2276
├── dec_3476
├── dec_4822
├── dec_6166
├── dec_6676
├── dec_931
├── dec_min_1802
├── dec_min_2924
└── dec_min_413
Example models directory structure#
/fefs/aswg/data/models/AllSky/20240131_allsky_v0.10.5_all_dec_base/
├── dec_2276
├── dec_3476
├── dec_4822
├── dec_6166
├── dec_6676
├── dec_931
├── dec_min_1802
├── dec_min_2924
└── dec_min_413
Example DL2 directory structure#
/fefs/aswg/data/mc/DL2/AllSky/20240131_allsky_v0.10.5_all_dec_base/
└── TestingDataset
├── dec_2276
│ ├── node_theta_10.0_az_102.199_
│ ...
│ └── node_theta_82.155_az_79.122_
├── dec_931
│ ├── node_theta_10.0_az_102.199_
...
│ └── node_theta_82.155_az_79.122_
├── dec_min_1802
│ ├── node_theta_10.0_az_102.199_
...
│ └── node_theta_82.155_az_79.122_
├── dec_min_2924
│ ├── node_theta_10.0_az_102.199_
...
│ ├── node_theta_82.155_az_271.199_
│ └── node_theta_82.155_az_79.122_
└── dec_min_413
├── node_theta_10.0_az_102.199_
...
├── node_theta_82.155_az_271.199_
└── node_theta_82.155_az_79.122_
Real Data analysis 💀#
Real data analysis is not supposed to be supported by these scripts. Use at your own risk.
Pipeline Support 🛠️#
So far the reference pipeline is lstchain.
There is however some support for ctapipe and hiperta as well (depending on lstmcpipe version).
The processing up to dl1 is relatively agnostic of the pipeline; working implementations exist for all of them.
In the case of hiperta a custom script converts the dl1 output to lstchain compatible files and the later stages
run using lstchain scripts.
In the case of ctapipe dl1 files can be produced using ctapipe-stage1. Once the dependency issues are solved and
ctapipe 0.12 is released, this will most likely switch to using ctapipe-process. We do not have plans to keep supporting older
versions longer than necessary currently.
Because the files are not compatible to lstchain and there is no support for higher datalevels in ctapipe yet, it is not possible
to use any of the following stages. This might change in the future.
Stages ⚙️#
After launching of the pipeline all selected tasks will be performed in order.
These are referred to as stages and are collected in lstmcpipe/stages.
Following is a short overview over each stage, that can be specified in the configuration.
r0_to_dl1
In this stage simtel-files are processed up to datalevel 1 and separated into files for training
and for testing.
For efficiency reasons files are processed in batches: N files (depending on paricle type
as that influences the averages duration of the processing) are submitted as one job in a jobarray.
To group the files together, the paths are saved in files that are passed to
python scripts in lstmcpipe/scripts which then call the selected pipelines
processing tool. These are:
lstchain: lstchain_mc_r0_to_dl1
ctapipe: ctapipe-stage1
rta: lstmcpipe_hiperta_r0_to_dl1lstchain (
lstmcpipe/hiperta/hiperta_r0_to_dl1lstchain.py)
dl1ab
As an alternative to the processing of simtel r0 files, existing dl1 files can be reprocessed.
This can be useful to apply different cleanings or alter the images by adding noise etc.
For this to work the old files have to contain images, i.e. they need to have been processed
using the no_image: False flag in the config.
The config key dl1_reference_id is used to determine the input files.
Its value needs to be the full prod_id including software versions (i.e. the name of the
directories directly above the dl1 files).
For lstchain the dl1ab script is used, ctapipe can use the same script as for simtel
processing. There is no support for hiperta!
merge_dl1
In this stage the previously created dl1 files are merged so that you end up with train and test datesets for the next stages.
train_test_split
Split the dataset into training and testing datasets, performing a random selection of files with the specified ratio (default=0.5).
train_pipe
IMPORTANT: From here on out only lstchain tools are available. More about that at the end.
In this stage the models to reconstruct the primary particles properties are trained
on the gamma-diffuse and proton train data.
At present this means that random forests are created using lstchains
lstchain_mc_trainpipe
Models will be stored in the models directory.
dl1_to_dl2
The previously trained models are evaluated on the merged dl1 files using lstchain_dl1_to_dl2 from
the lstchain package.
DL2 data can be found in DL2 directory.
dl2_to_irfs
Point-like IRFs are produced for each set of offset gammas.
The processing is performed by calling lstchain_create_irf_files.
dl2_to_sensitivity
A sensitivity curve is estimated using a script based on pyirf which performs a cut optimisation
similar to EventDisplay.
The script can be found in lstmcpipe/scripts/script_dl2_to_sensitivity.py.
This does not use the IRFs and cuts computed in dl2_to_irfs, so this can not be compared to observed data.
It is a mere benchmark for the pipeline.
📈 Logs and data output#
Job logs are stored along with the produced data for each stage. E.g. in
$PPROD_ID/TrainingDataset/dec_2276/Protons/node_theta_16.087_az_108.090_/job_logs_r0dl1/
Higher-level, lstmcpipe logs are produced and stored in the $HOME/LSTMCPIPE_PROD_LOGS/ directory created when installing lstmcpipe.
Every time a full MC production is launched, two files with logging information are created:
log_reduced_Prod{3,5}_{PROD_ID}.ymllog_onsite_mc_r0_to_dl3_Prod{3,5}_{PROD_ID}.yml
The first one contains a reduced summary of all the scheduled job ids (to which particle the job corresponds to),
while the second one contains the same plus all the commands passed to slurm.